Get Rational! From Drug Discovery to Green Chemistry
An informal literature mini review:
Robert E Babine
Rebexsess Discovery Chemistry
published on www.rebexsess.com, March 2013
The Next Wave of Protein Structure Based
Design
Upon REB’s arrival at Agouron Pharmaceuticals 20 years ago he received a light blue T-shirt with the Agouron logo and the words “Get Rational” on the front. At that time it was approximately a decade into the use of protein structure as a tool for drug discovery. In the late 1990’s the fruits of that pioneering technology were beginning to see the light with several HIV protease inhibitors, from startups like Agouron and Vertex as well as big pharma, becoming marketed drugs. These protease inhibitors are key components of life saving drug cocktails that have halted the spread of HIV and its progression into AIDS. The rational drug design paradigm was a big success and the tools and strategies of Structure Based Drug Design (SBDD) are now incorporated into many drug discovery programs.
In SBDD the structures of key ligands bound to the protein (enzyme or receptor) provide key data in advancing the SAR of a discovery program. In the rational design of enzyme inhibitors as drugs, detailed information about enzyme mechanism and protein structure are merged together with principles of Medicinal Chemistry to create novel and effective small molecules. The orally bioavailable anti Hepatitis C drugs Telaprevir (Incivek) and Boceprevir (Victrelis) were rationally invented and are now saving lives. (See the Rebexsess June 2011 mini review for discussion)
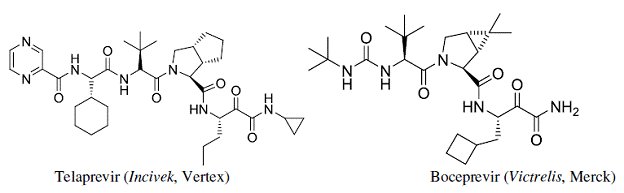
Rebexsess offers key SBDD services (See the Rebexsess June 2011 mini review for discussion)
The Next Frontier: Structure Based Enzyme
Engineering (SBEE)
The Vertex process chemistry to produce Telaprevir involves as a key step the chemical desymmetrization of the meso intermediate 1, to produce 2 or a synthetic equivalent. An analogous approach towards the synthesis of Boceprevir occurs via intermediate 3.
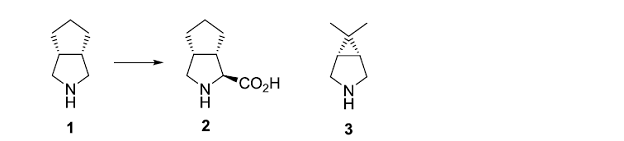
• G. J. Tanoury, M. Chen, and J. E. Cochran, US Pat.,
US20070087973, Vertex Pharmaceuticals Inc., 2007
Two recent reports have described the use of novel,
engineered enzymes to carry out the oxidative desymmetrization of both 1 and 3. These efficient,
chemoenzymatic processes for manufacture of these intermediates uses an enzyme
discovered through directed evolution, the N336S/I246M mutant of the
FAD-containing monoamine oxidase from Aspergillus
• Znabet, A.; Polak, M. M.; Janssen, E.; de Kanter, F. J.;
Turner, N. J.; Orru, R. V.; Ruijter, E.: A highly efficient synthesis of
telaprevir by strategic use of biocatalysis and multicomponent reactions. Chem Commun (Camb) 2010, 46, 7918-20; doi= 10.1039/c0cc02823a.
• Li, T.; Liang, J.; Ambrogelly, A.; Brennan, T.; Gloor, G.;
Huisman, G.; Lalonde, J.; Lekhal, A.; Mijts, B.; Muley, S.; Newman, L.; Tobin,
M.; Wong, G.; Zaks, A.; Zhang, X.: Efficient, chemoenzymatic process for
manufacture of the boceprevir bicyclic [3.1.0]proline intermediate based on
amine oxidase-catalyzed desymmetrization. J
Am Chem Soc 2012, 134, 6467-72; doi= 10.1021/ja3010495.
Over the last decade, scientific and technological advances have established biocatalysis as a practical and environmentally friendly alternative to traditional metallo- and organocatalysis in chemical synthesis, both in the laboratory and on an industrial scale. Key advances in DNA sequencing and gene synthesis are at the base of tremendous progress in tailoring biocatalysts by protein engineering and design, and the ability to reorganize enzymes into new biosynthetic pathways. These mutagenesis based tools, referred to as directed evolution, harness the power of natural selection to evolve proteins with desirable properties not found in nature.
These engineering methods are now in use and have enormous potential in “green chemistry” to produce new catalyst useful for:
- Biofuels
- Industrial Synthesis of commodity organic chemicals
- Industrial Synthesis of “high values” chemicals such as pharmaceutical intermediates
• Bornscheuer, U. T.; Huisman, G. W.; Kazlauskas,
R. J.; Lutz, S.; Moore, J. C.; Robins, K.: Engineering the third wave of
biocatalysis. Nature 2012, 485, 185-94; doi=
10.1038/nature11117.
• Lutz, S.: Beyond directed
evolution--semi-rational protein engineering and design. Curr Opin Biotechnol 2010,
21, 734-43; doi= 10.1016/j.copbio.2010.08.011.
• Quin, M. B.; Schmidt-Dannert, C.:
Engineering of biocatalysts - from evolution to creation. ACS Catal 2011, 1, 1017-1021; doi= 10.1021/cs200217t.
Directed evolution is a key tool for enzyme engineering; however, multiple random mutations rapidly lead to a numbers problem where too many mutant proteins exist to be evaluated in a timely manner. Hence there is a crucial need for intelligent selection of targeted mutations. The use of protein structure, coupled with computational models for enzyme function, allow one to rationally tackle the numbers problem. Both SBDD and SBEE use related computational tools, in SBDD models based on protein-ligand complexes are used to design new and better ligands while in SBEE, models based on protein-ligand complexes are used to design better enzymes. Thus, the same key tools and intuition important for using protein structure in drug discovery apply to research in biocatalysis. Rebexsess is well positioned to rationally tackle the numbers problems in biocatalyst development.
A few examples from the literature are discussed below to emphasize how protein structure has been utilized in biocatalyst creation and the vast potential for use of SBEE in future endeavors.
Directed evolution of stereoselective and
thermostable enzymes
The research group headed by Manfred T. Reetz has been very active in development of new enzymes for key organic transformations. They have developed methodology and protocols to address the numbers problem in directed evolution. The use of protein structure plays a significant role in library design, especially in identifying pairs of residues to be mutated simultaneously. In their research they have discovered new enzymes for important transformations. A select few are illustrated below:
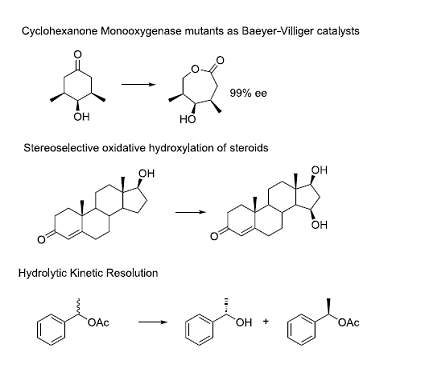
• Wu, Q.; Soni, P.; Reetz, M. T.:
Laboratory Evolution of Enantiocomplementary Candida
• Informative Reetz website: www.kofo.mpg.de/en/research/organic-synthesis
Sitagliptin:
Biocatalysis in Drug Manufacturing
In an elegant study, an efficient biocatalytic process was developed, to replace a rhodium-catalyzed asymmetric enamine hydrogenation, for the large-scale manufacture of the antidiabetic compound sitagliptin. The starting point was an enzyme that had the catalytic machinery to perform the desired chemistry but lacked any activity toward the prositagliptin substrate ketone. Initial studies used protein structure and computational model building of an enzyme-substrate complex to design initial mutant proteins. A transaminase enzyme with marginal activity for the synthesis of the appropriate chiral amine was discovered. This mutant enzyme was then further engineered via several rounds of directed evolution for practical application in a manufacturing setting. The resultant biocatalysts showed broad applicability toward the synthesis of chiral amines that previously were accessible only via resolution. This work underscores the maturation of biocatalysis to enable efficient, economical, and environmentally benign processes for the manufacture of pharmaceuticals.
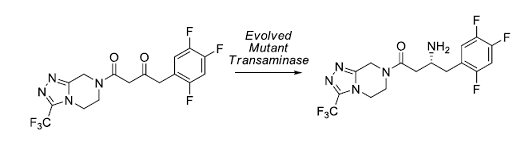
• Savile, C.
K.; Janey, J. M.; Mundorff, E. C.; Moore, J. C.; Tam, S.; Jarvis, W. R.;
Colbeck, J. C.; Krebber, A.; Fleitz, F. J.; Brands, J.; Devine, P. N.; Huisman,
G. W.; Hughes, G. J.: Biocatalytic asymmetric synthesis of chiral amines from
ketones applied to sitagliptin manufacture. Science
2010, 329, 305-9; doi= 10.1126/science.1188934.
Terpenoid Cyclases
The terpenoid cyclase class of enzymes catalyze reactions that can create highly complex organic compounds from simple acyclic starting materials. The enzyme taxadiene synthase converts the acyclic geranylgeranyl pyrophosphate (4) into the complex tricyclic hydrocarbon taxadiene (5) in a single step! Taxadiene is the biosynthetic precursor for the antitumor agent taxol (6); the types of oxidative enzymes involved in the conversion of 5 into 6 are also fascinating and are of potential value in biocatalyst. The terpenoid cyclase class of enzymes has been well studied from a mechanistic and structural point of view.
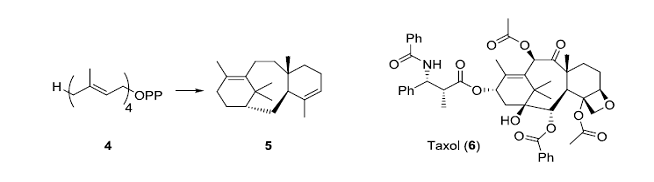
• Cane, D.
E.: Enzymatic Formation of Sesquiterpenes. Chem
Rev 1990, 90, 1089-1103; doi=10.1021/cr00105a002.
• Christianson,
D. W.: Structural biology and chemistry of the terpenoid cyclases. Chem Rev 2006, 106, 3412-42; doi= 10.1021/cr050286w.
A practical limitation in the use of terpenoid cyclases in
biocatalyst is that the acyclic pyrophosphate starting materials are not
readily available as starting materials.
A significant advance in using this class of enzymes involves using
organisms engineered to overproduce both the terpenoid cyclase enzyme and the
synthesis (from acetate) of its starting material. In key work from the Keasling lab, funded by
the Gates Foundation, a synthetic precursor (9) of the important anti-malarial agent Artemisinin
(10) is biosynthesized in engineered
organisms. In the box below are the key
biosynthetic steps; conversion of acetate into farnesyl pyrophosphate (7), terpenoid cyclase conversion of 7 into 8, followed by three oxidative steps to produce 9.
The product of this synthetic biology pathway (9) can be converted into Artemisinin (10) by organic synthesis.
This work makes a key anti-malarial compound available to save lives.
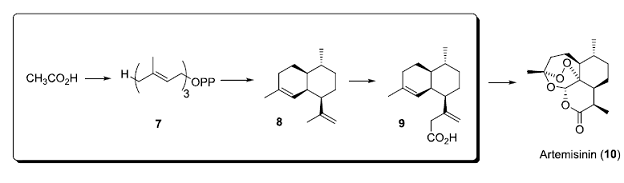
• Chang, M. C.; Keasling, J. D.:
Production of isoprenoid pharmaceuticals by engineered microbes. Nat Chem Biol 2006, 2, 674-81; doi= 10.1038/nchembio836.
The diversity and complexity of
products generated by terpenoid cyclase enzymes, coupled with the ability to
incorporate these enzymes into engineered organisms suggest the future
importance of the engineering of novel enzymes to produce new important
chemicals as commodity products, drug intermediates and biofuels. In a significant study, Cane and Christianson
have shown that targeted point mutations in the terpenoid cyclase enzyme epi-isozizaene synthase can significantly alter
the product distribution. The wild type
enzyme produces 11 as the major
product (80% of mixture) with 13 as
a minor product (2%); the F198A mutant does not produce 11, but rather 12 as the
major product (24%) with 13 as a
significant component (20%) of the mixture.
This work demonstrates the use crystal structures in the design of key
mutations and the ability of this class of enzyme to be converted into novel
catalysts for new products.
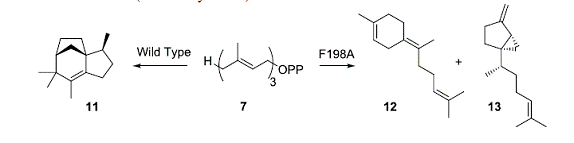
• Aaron, J. A.; Lin, X.; Cane, D. E.;
Christianson, D. W.: Structure of epi-isozizaene synthase from Streptomyces
coelicolor A3(2), a platform for new terpenoid cyclization templates. Biochemistry 2010, 49, 1787-97; doi= 10.1021/bi902088z.
In another dramatic example, the Keasling group performed directed evolution on the terpenoid cyclase enzyme g-humulene synthase. This highly promiscuous enzyme produces 52 different isomeric products from the same substrate, farnesyl pyrophosphate (7), with g-humulene (14) as the major product (45% of complex mixture). Using a homology model of this enzyme as a starting point several new enzymes were created using directed evolution. One enzyme, a triple mutant, produced 14 as 85% of the mixture. Another enzyme, a point mutant, produces the isomeric farnesenes 15 (54%) and 16 (34%); these products are precursors of the important biofuel farnesane (17). Two other triple mutants produce 18 and 19 respectively as major components (~80%) of the mixture. Clearly the terpenoid cyclase class of enzymes lend themselves to redesign and many crystal structures are available for new SBEE applications.
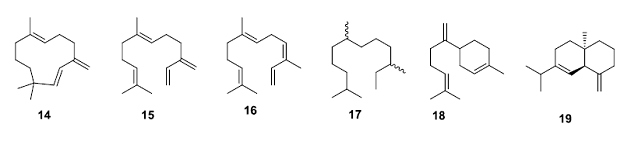
• Yoshikuni,
Y.; Ferrin, T. E.; Keasling, J. D.: Designed divergent evolution of enzyme
function. Nature 2006, 440, 1078-82; doi= 10.1038/nature04607.
Rebexsess approach to SBEE
The Rebexsess approach to engineering new enzymes involves using a rational approach to solving the numbers problem in directed evolution, structure based enzyme engineering (SBEE). The use of protein structure, coupled with computational models for enzyme function, allow one to rationally tackle the numbers problem. Both SBDD and SBEE use related computational tools, in SBDD models based on protein-ligand complexes are used to design new and better ligands while in SBEE, models based on protein-ligand complexes are used to design better enzymes. Thus, the same key tools and intuition important for using protein structure in drug discovery apply to research in biocatalysis. Rebexsess expertise in mechanistic organic chemistry and in understanding the molecular recognition of protein-ligand complexes is ideally suited for building high quality 3D models for substrates and intermediates along the reaction pathway. These 3D models and homology in a family of enzymes, then lead to candidates for mutation and narrowing down the residues to be mutated to. By taking into account key residues and neighboring enzyme residues (in 3D space) rational focused libraries for Round 1 in a directed evolution scheme can be designed.
The major benefit from SBEE is a well designed library of mutations for a focused directed evolution that will get better coverage than a significantly larger library of more random mutants. Rebexsess can deliver to clients rationally designed mutants for getting good coverage of enzyme residues for input into directed evolution experiments. These libraries would allow for the screening of substantially fewer mutants than random libraries, thus being cost effective.
Rebexsess Core Technology
Rebexsess Core Technology for an enzyme family involves the integration of protein structure and protein homology with mechanistic organic chemistry to analyze and design mutant proteins for site directed mutagenesis in directed evolution. Rapid screening of mutants and detailed evaluation of interesting mutants generates new data that adds to Rebexsess core technology for this family of enzymes. This information is incorporated into this next cycle of analysis and design; thus, making this an iterative cycle.
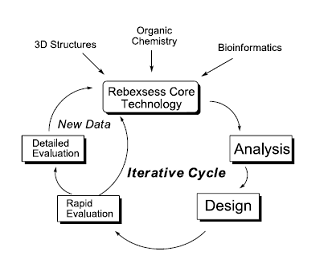
The intellectual challenge in developing Rebexsess Core Technology is to be able to effectively integrate biochemical and protein structural data with the complex and often perplexing organic reaction mechanisms possibly involving intermediates and produce efficient solutions. With expertise in both organic chemistry and protein structure Rebexsess is uniquely positioned to work effectively at this integrated multidisciplinary interface.
The Rebexsess Core Technology identifies key targets (product and enzyme), key enzymes for
mutagenesis, along with 3D alignments.
Key mechanistic pathways are built as 3D models aligned together based
on structural, sequence and homology alignments of key enzymes. If necessary 3D homology models for key
enzymes and transformations are built.
The Rebexsess Core Technology develops coherent 3D models for different enzymatic transformations and
possible roles for key residues. These
alignments allow for the intelligent design of new enzymes by suggesting
residues that have a conserved, though obscure function and residues that would
likely be targets of either specific or non-specific mutations. Hence this technology allows for SBEE, the
computation analysis, rational design, directed evolution, rapid evaluation of
mutant enzymes, and detailed enzymatic analyses of key enzymes in an iterative
fashion.
Rebexsess offers the
following SBEE Services:
(1)
As a key component of the analysis stage of the Rebexsess SBEE core technology the first step involves development of high quality 3D models of the reaction pathway catalyzed by the enzyme of interest. This is a crucial step since it allows one to visualize and understand how the enzyme catalyzes the reaction and it allows for identification of key protein residues and their potential role in catalysis. This forms the basis for the rational identification of putative key protein residues involved in catalysis and the prioritization of targeted point mutations and key pairs of residues close in 3D space.
As an illustration, the single step conversion of the acyclic geranylgeranyl pyrophosphate (4) starting material into the complex tricyclic hydrocarbon taxadiene (5) catalyzed by was modeled. The starting points for this exercise were the crystal structure of taxadiene synthase from Pacific Yew in complex with Mg2+ and 2-fluorogeranylgeranyl diphosphate (PDB entry 3P5R; also see 3P5P) and the detailed mechanistic (labeling) and quantum mechanical studies on this fascinating conversion. As shown in the scheme below, ionization of 4 (loss of pyrophosphate) followed by a number of carbocation rearrangements and proton loss lead to 5. Effective model building must take into account an effective fit into the active site as well results from mechanistic labeling studies and the chemical reactivity of the substrate and reactive intermediates. As a model system, Rebexsess has built 3D models for this enzyme catalyzed reaction, consistent with experimental data and this model suggests the role of the enzyme in exerting conformational control of substrate folding, binding of carbocation intermediates and identification of the pyrophosphate leaving group as the base in the final deprotonation step in this sequence.
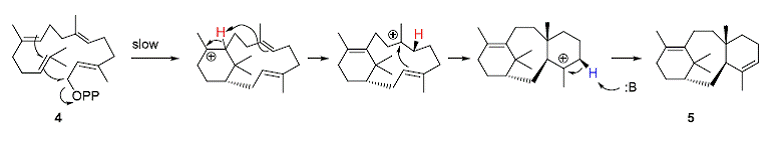
• Koksal,
M.; Jin, Y.; Coates, R. M.; Croteau, R.; Christianson, D. W.: Taxadiene
synthase structure and evolution of modular architecture in terpene
biosynthesis. Nature 2011, 469, 116-20; doi=
10.1038/nature09628.
• Hong, Y. J.; Tantillo, D. J.: The
taxadiene-forming carbocation cascade. J
Am Chem Soc 2011, 133, 18249-56; doi= 10.1021/ja2055929.
(2) Analysis and Design
The Rebexsess advantage is the ability to couple diverse detailed information about complex organic reactions with the fundamentals of the molecular recognition of protein-ligand complexes to generate models useful for the design of target mutations. The use Rebexsess high quality computational models for enzyme function, allow one to rationally tackle the numbers problem in directed evolution.
These models allow us to identify key enzyme residues, their possible role in catalysis, homologous residues in related enzymes, and residues that are close in 3D space. This information can then be used to design libraries of mutant proteins as input for focused directed evolution.
(3) Assay Design
Rebexsess provides consulting services that can assist with the design of key assays. In addition, Rebexsess can design novel substrates that allow for rapid screening. Using a paradigm established during the optimization of an enzymatic step in the process to produce the pharmaceutical sitagliptin screening conditions should be rendered more stringent with more favorable product mixtures and rising activity. Rebexsess can provide consulting to assist in assay conditions designed to detect new mutants with properties appropriate for the level of optimization needed for that round.
(4) Iterative Process
The key for using protein structure in any design process is to incorporate experimental data from initial rounds into new and better models. Thus, an iterative process with several rounds of modeling, design and experimental feedback, is most efficient. The first round in this process begins with evaluation of current protein structures of the relevant enzymes and interpreting detailed mechanistic studies in the context of these proteins. This is used to build the initial 3D models and design initial mutant libraries. In the next round, upon analysis of the screening data and biochemical data from key mutant enzymes models will are refined as warranted by the data. This new model is then used to assist in the design of mutant libraries for the next directed evolution cycle. This comprises the Rebexsess Core Technology.